Cell Differentation
Research Lines
-Role of SUMO in cell viability and differentiation
-BET proteins function in cell proliferation and differentiation
Introduction
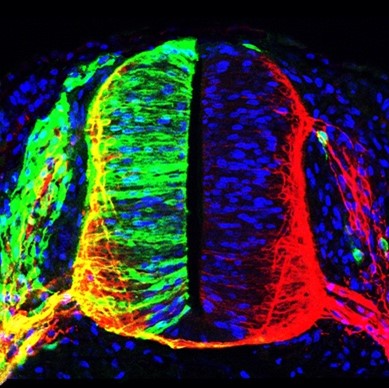 Today it is clear that human pathologies such as cancer, degenerative diseases and developmental disorders do not have simple and common origins. The diversity of factors that lead to the alteration of homeostasis in each type of tissue requires a comprehensive analysis of the exact causes underlying a particular disease to efficiently apply appropriate intervention therapies. This is closely linked to very current concepts such as precision and personalized medicine. In this context, our main research objective focuses on deciphering the molecular mechanisms that control cell viability and differentiation, especially in the vertebrate nervous system. In particular, we study the post-translational modification of proteins by covalent attachment of the SUMO polypeptide (sumoylation), and also chromatin adapters of the BET family, which are notably linked to cancer. We are keenly interested in cell signaling, transcriptional control, and chromatin-associated factors and architecture, to define novel and suitable targets and pathways for precise and effective therapeutic intervention.
Today it is clear that human pathologies such as cancer, degenerative diseases and developmental disorders do not have simple and common origins. The diversity of factors that lead to the alteration of homeostasis in each type of tissue requires a comprehensive analysis of the exact causes underlying a particular disease to efficiently apply appropriate intervention therapies. This is closely linked to very current concepts such as precision and personalized medicine. In this context, our main research objective focuses on deciphering the molecular mechanisms that control cell viability and differentiation, especially in the vertebrate nervous system. In particular, we study the post-translational modification of proteins by covalent attachment of the SUMO polypeptide (sumoylation), and also chromatin adapters of the BET family, which are notably linked to cancer. We are keenly interested in cell signaling, transcriptional control, and chromatin-associated factors and architecture, to define novel and suitable targets and pathways for precise and effective therapeutic intervention.
Role of SUMO in cell viability and differentiation
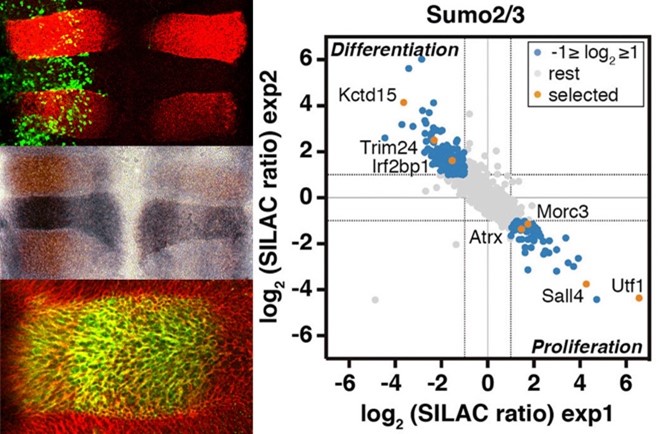 SUMO (Small Ubiquitin-like MOdifier) is a small polypeptide, similar to ubiquitin, able to covalently attach to proteins as a post-translational modifier. SUMO is essential in eukaryotes and participates in the regulation of almost all cellular processes. Sumoylation is involved in transcriptional control, nucleocytoplasmic transport, protein and genome stability, and enzymatic activity modulation. The process of modification involves SUMO transfer from the conjugating enzyme UBC9 to the target protein, frequently mediated by SUMO ligases, which favor and enhance transfer and are key regulators of the process. Among the most studied SUMO ligases are those of the PIAS family. Specific SUMO proteases of the SENP family are also key regulators of the process since they detach SUMO from targets. Knowing in detail SUMO targets and components of the SUMO pathway involved in regulating proliferation, differentiation and cell viability, together with the associated regulatory mechanisms, is of great therapeutic interest for developmental and nervous system disorders, and cancer. We have discovered that the transcription factor KROX20 displays SUMO ligase activity on its NAB corepressors during hindbrain development. We have identified the SUMO protease SENP7 as an essential factor for neurogenesis and as a cell viability-promoting factor in tumor cells under oxygen and glucose-limiting conditions, being a prognostic marker for colon cancer. Through a SILAC-based proteomic study, we have identified differentially sumoylated proteins during the process of neurogenesis. Among these, we have described the transcription factor UTF1 as a new SUMO target, the sumoylation of which regulates its affinity for the chromatin and the recruitment of the decapping enzyme DCP1A for transcriptional control of bivalent developmental genes. In another proteomic study, we have identified more than one hundred proteins sumoylated in response to oxygen and glucose deprivation, which are conditions associated with ischemia and also frequent inside solid tumors, and we have studied how the sumoylation of specific factors impacts cell viability under these deleterious conditions. We continue research on SUMO targets and SUMO ligases and proteases linked to several pathologies.
SUMO (Small Ubiquitin-like MOdifier) is a small polypeptide, similar to ubiquitin, able to covalently attach to proteins as a post-translational modifier. SUMO is essential in eukaryotes and participates in the regulation of almost all cellular processes. Sumoylation is involved in transcriptional control, nucleocytoplasmic transport, protein and genome stability, and enzymatic activity modulation. The process of modification involves SUMO transfer from the conjugating enzyme UBC9 to the target protein, frequently mediated by SUMO ligases, which favor and enhance transfer and are key regulators of the process. Among the most studied SUMO ligases are those of the PIAS family. Specific SUMO proteases of the SENP family are also key regulators of the process since they detach SUMO from targets. Knowing in detail SUMO targets and components of the SUMO pathway involved in regulating proliferation, differentiation and cell viability, together with the associated regulatory mechanisms, is of great therapeutic interest for developmental and nervous system disorders, and cancer. We have discovered that the transcription factor KROX20 displays SUMO ligase activity on its NAB corepressors during hindbrain development. We have identified the SUMO protease SENP7 as an essential factor for neurogenesis and as a cell viability-promoting factor in tumor cells under oxygen and glucose-limiting conditions, being a prognostic marker for colon cancer. Through a SILAC-based proteomic study, we have identified differentially sumoylated proteins during the process of neurogenesis. Among these, we have described the transcription factor UTF1 as a new SUMO target, the sumoylation of which regulates its affinity for the chromatin and the recruitment of the decapping enzyme DCP1A for transcriptional control of bivalent developmental genes. In another proteomic study, we have identified more than one hundred proteins sumoylated in response to oxygen and glucose deprivation, which are conditions associated with ischemia and also frequent inside solid tumors, and we have studied how the sumoylation of specific factors impacts cell viability under these deleterious conditions. We continue research on SUMO targets and SUMO ligases and proteases linked to several pathologies.
BET proteins function in cell proliferation and differentiation
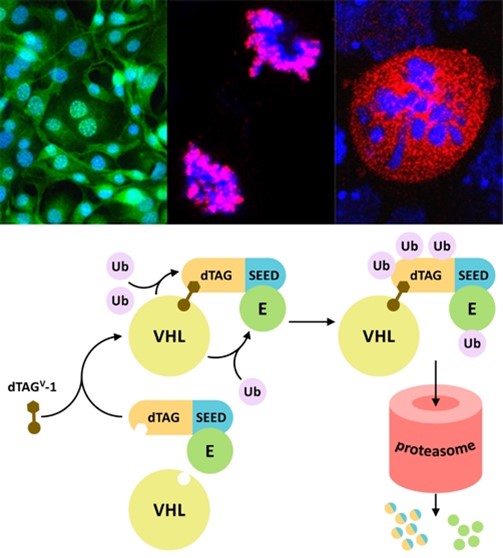 BET proteins (BRD2, BRD3, BRD4 and BRDT) are characterized by the presence of two bromodomains able to recognize acetylated histones in the chromatin and an extra-terminal (ET) domain, exclusive to this family. These proteins are essential for development, play a prominent role in cell cycle progression, but also in differentiation, and are frequently dysregulated in most cancer types. We investigate BET proteins in the context of cell proliferation and differentiation and related to disease. Our results are of interest for cancer and other diseases, particularly regarding drug discovery, cell therapy and regenerative medicine. We have identified an essential dimerization domain in BET proteins, and described the Pleiotrophin antagonism on BRD2 during neurogenesis. We have assigned a role to the LYAR-BRD2 interaction in controlling the expression of the pluripotency factor Nanog after inducing differentiation. We have shown that NIPBL, the cohesin loading factor associated with the Cornelia de Lange syndrome (CdLS), interacts with the ET domain of BRD4 to regulate the expression of developmental genes, explaining why mutations in BRD4 show a CdLS-like phenotype. Recently, we have also characterized the interaction of BET members with the envelope (E) protein of the SARS-CoV-2 virus, opening new therapeutic approaches to fight the COVID-19 disease. Thus, our current research on BET proteins is related to different pathologies, such as cancer, CdLS and COVID-19, with special interest in uncovering new approaches for efficient BET inhibition.
BET proteins (BRD2, BRD3, BRD4 and BRDT) are characterized by the presence of two bromodomains able to recognize acetylated histones in the chromatin and an extra-terminal (ET) domain, exclusive to this family. These proteins are essential for development, play a prominent role in cell cycle progression, but also in differentiation, and are frequently dysregulated in most cancer types. We investigate BET proteins in the context of cell proliferation and differentiation and related to disease. Our results are of interest for cancer and other diseases, particularly regarding drug discovery, cell therapy and regenerative medicine. We have identified an essential dimerization domain in BET proteins, and described the Pleiotrophin antagonism on BRD2 during neurogenesis. We have assigned a role to the LYAR-BRD2 interaction in controlling the expression of the pluripotency factor Nanog after inducing differentiation. We have shown that NIPBL, the cohesin loading factor associated with the Cornelia de Lange syndrome (CdLS), interacts with the ET domain of BRD4 to regulate the expression of developmental genes, explaining why mutations in BRD4 show a CdLS-like phenotype. Recently, we have also characterized the interaction of BET members with the envelope (E) protein of the SARS-CoV-2 virus, opening new therapeutic approaches to fight the COVID-19 disease. Thus, our current research on BET proteins is related to different pathologies, such as cancer, CdLS and COVID-19, with special interest in uncovering new approaches for efficient BET inhibition.
Funding:
– AEI (PID, PGC)
– Junta de Andalucía
– CSIC
Lara-Ureña N, Gómez-Marín E, Pozuelo-Sánchez I, Reyes JC, García-Domínguez M (2024) SARS-CoV-2 E protein interacts with BRD2 and BRD4 SEED domains and alters transcription in a different way than BET inhibition. Cell Mol Life Sci 81: 313.
Samra N, Jansen NS, Morani I, Kakun RR, Zaid R, Paperna T, García-Domínguez M, Viner Y, Frankenthal H, Shinwell ES, Portnov I, Bakry D, Shalata A, Shapira Rootman M, Kidron D, Claessens LA, Wevers RA, Mandel H, Vertegaal ACO, Weiss K (2023) Exome sequencing links the SUMO protease SENP7 with fatal arthrogryposis multiplex congenita, early respiratory failure and neutropenia. J Med Genet 60: 1133-1141.
Gallardo-Chamizo F, Lara-Ureña N, Correa-Vázquez JF, Reyes JC, Gauthier BR, García-Domínguez M (2022) SENP7 overexpression protects cancer cells from oxygen and glucose deprivation and associated with poor prognosis in colon cancer. Genes Dis 9: 1419-1422.
Lara-Ureña N, Jafari V, García-Domínguez M (2022) Cancer-Associated Dysregulation of Sumo Regulators: Proteases and Ligases. Int J Mol Sci 23: 8012.
García-Gutiérrez P, García-Domínguez M (2021) SUMO control of nervous system development. Semin Cell Dev Biol 132: 203-212.
Correa-Vázquez JF, Juárez-Vicente F, García-Gutiérrez P, Barysch SV, Melchior F, García-Domínguez M (2021) The Sumo proteome of proliferating and neuronal-differentiating cells reveals Utf1 among key Sumo targets involved in neurogenesis. Cell Death Dis 12: 305.
Luna-Peláez N, March-Díaz R, Ceballos-Chávez M, Guerrero-Martínez JA, Grazioli P, García-Gutiérrez P, Vaccari T, Massa V, Reyes JC & García-Domínguez M (2019) The Cornelia de Lange Syndrome-associated factor NIPBL interacts with BRD4 ET domain for transcription control of a common set of genes. Cell Death Dis 10: 548.
Luna-Peláez N & García-Domínguez M (2018) Lyar-Mediated recruitment of Brd2 to the chromatin attenuates Nanog downregulation following induction of differentiation. J Mol Biol 430: 1084-1097.
Juárez-Vicente F, Luna-Peláez N & García-Domínguez M (2016) The Sumo protease Senp7 is required for proper neuronal differentiation. BBA Mol Cell Res 1863: 1490-1498.
García-Gutiérrez P, Juárez-Vicente F, Wolgemuth DJ & García-Domínguez M (2014) Pleiotrophin antagonizes Bromodomain-containing protein 2 (Brd2) during neuronal differentiation. J Cell Sci 127: 2554-2564.
Alfonso-Pérez T, Domínguez-Sánchez MS, García-Domínguez M & Reyes JC (2014) Cytoplasmic interaction of the tumor supresor protein hSNF5 with dynamin-2 controls endocitosis. Oncogene 33: 3064-3074.
Ceballos-Chávez M, Rivero S, García-Gutiérrez P, Rodríguez-Paredes M, García-Domínguez M, Bhattacharya SS & Reyes JC (2012) Control of neuronal differentiation by Braf35 sumoylation. Proc Natl Acad Sci USA 109: 8085-8090.
García-Gutiérrez P, Mundi M, & García-Domínguez M (2012) Association of bromodomain BET proteins to the chromatin requires dimerization through the conserved motif B. J Cell Sci 125: 3671–3680.
García-Gutiérrez P, Juárez-Vicente F, Gallardo-Chamizo F, Charnay P & García-Domínguez M (2011) The transcription factor Krox20 is an E3 ligase that sumoylates its Nab coregulators. EMBO Rep 12: 1018-23.








